The coronavirusbrazil package provides a tidy format dataset of the 2019 Novel Coronavirus COVID-19 (2019-nCoV) epidemic for Brazil. The datasets were obtained from Ministerio da Saúde, brasil.io and Secretaria de Saúde - RJ.
This repository was inspired by the RamiKrispin/coronavirus package repository.
Installation
You can install the released version of coronavirusbrazil from CRAN with:
# install.packages("devtools") devtools::install_github("mralbu/coronavirusbrazil")
Usage
The package contains the following datasets:
library(coronavirusbrazil)
library(ggplot2)
data("coronavirus_br")
head(coronavirus_br)
#> # A tibble: 6 x 10
#> date cases deaths new_cases new_deaths death_rate percent_case_in~
#>
#> 1 2020-02-25 0 0 NA NA NaN NA
#> 2 2020-02-26 1 0 1 0 0 Inf
#> 3 2020-02-27 1 0 0 0 0 0
#> 4 2020-02-28 1 0 0 0 0 0
#> 5 2020-02-29 2 0 1 0 0 100
#> 6 2020-03-01 2 0 0 0 0 0
#> # ... with 3 more variables: percent_death_increase , days_gt_10 ,
#> # days_gt_100 plot_coronavirus(coronavirus_br, xaxis = "date", yaxis = "cases", log_scale = F, linear_smooth = F)
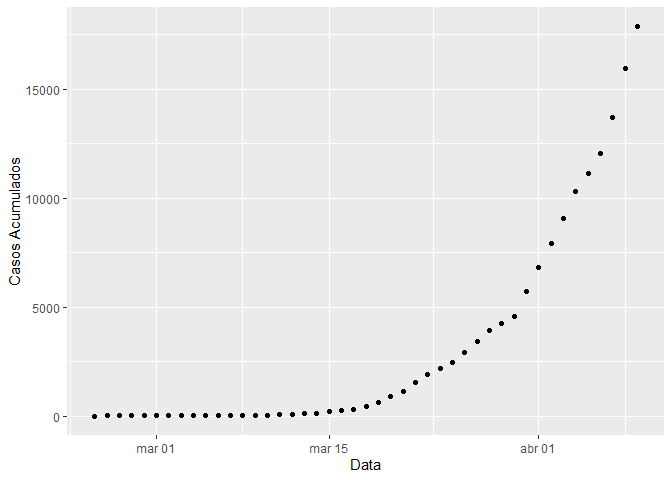
data("coronavirus_br_states")
head(coronavirus_br_states)
#> # A tibble: 6 x 11
#> # Groups: state [1]
#> state date cases deaths new_cases new_deaths death_rate percent_case_in~
#>
#> 1 RO 2020-02-25 0 0 0 0 NaN NaN
#> 2 RO 2020-02-26 0 0 0 0 NaN NaN
#> 3 RO 2020-02-27 0 0 0 0 NaN NaN
#> 4 RO 2020-02-28 0 0 0 0 NaN NaN
#> 5 RO 2020-02-29 0 0 0 0 NaN NaN
#> 6 RO 2020-03-01 0 0 0 0 NaN NaN
#> # ... with 3 more variables: percent_death_increase , days_gt_10 ,
#> # days_gt_100 plot_coronavirus(coronavirus_br_states, yaxis = "percent_case_increase", color = "state", filter_variable = "state", facet = "state", filter_values = c("RJ", "SP", "DF", "CE", "RS", "MG"), log_scale = TRUE, linear_smooth = TRUE)
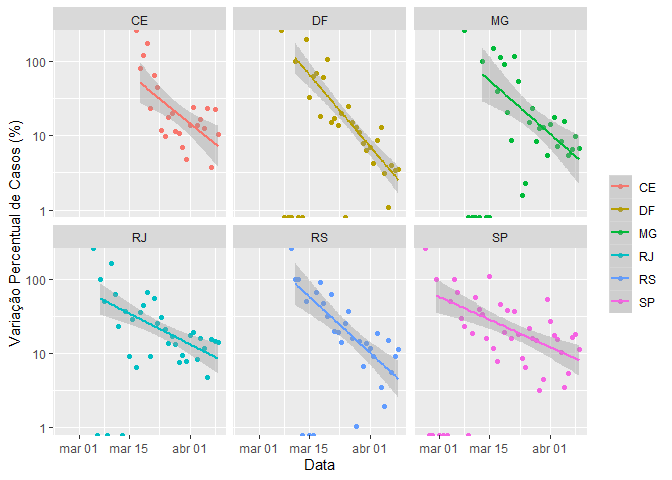
data("coronavirus_br_cities")
head(coronavirus_br_cities)
#> # A tibble: 6 x 17
#> # Groups: city [1]
#> city date state place_type cases deaths is_last estimated_popul~
#>
#> 1 Abae~ 2020-03-31 PA city 1 0 FALSE 157698
#> 2 Abae~ 2020-04-01 PA city 1 0 FALSE 157698
#> 3 Abae~ 2020-04-02 PA city 1 0 FALSE 157698
#> 4 Abae~ 2020-04-03 PA city 1 0 FALSE 157698
#> 5 Abae~ 2020-04-04 PA city 1 0 FALSE 157698
#> 6 Abae~ 2020-04-05 PA city 1 0 FALSE 157698
#> # ... with 9 more variables: city_ibge_code ,
#> # confirmed_per_100k_inhabitants , death_rate , new_cases ,
#> # new_deaths , percent_case_increase ,
#> # percent_death_increase , days_gt_10 , days_gt_100 There are also geospatial datasets avaiable:
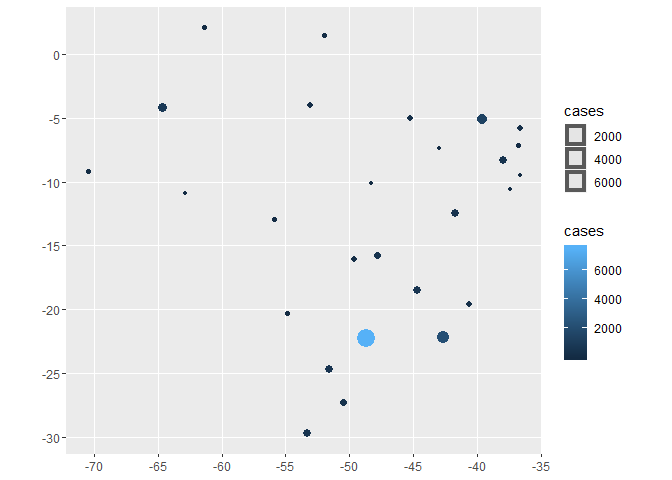
dplyr::glimpse(spatial_br_states) #> Observations: 27 #> Variables: 16 #> $ id"AC", "AL", "AM", "AP", "BA", "CE", "DF", "E... #> $ name "Acre", "Alagoas", "Amazonas", "Amapá", "Bah... #> $ uf "AC", "AL", "AM", "AP", "BA", "CE", "DF", "E... #> $ codigo 12, 27, 13, 16, 29, 23, 53, 32, 52, 21, 31, ... #> $ regiao "Norte", "Nordeste", "Norte", "Norte", "Nord... #> $ geometry [, $ date
2020-04-09, 2020-04-09, 2020-04-09, 2020-04... #> $ cases 62, 37, 899, 128, 559, 1425, 527, 273, 179, ... #> $ deaths 2, 3, 40, 2, 19, 55, 13, 6, 7, 12, 15, 2, 2,... #> $ new_cases 8, 0, 95, 21, 62, 134, 18, 46, 21, 43, 41, 4... #> $ new_deaths 0, 1, 10, 0, 4, 12, 1, 0, 0, 1, 1, 0, 1, 1, ... #> $ death_rate 0.03225806, 0.08108108, 0.04449388, 0.015625... #> $ percent_case_increase 14.8148148, 0.0000000, 11.8159204, 19.626168... #> $ percent_death_increase 0.000000, 50.000000, 33.333333, 0.000000, 26... #> $ log_cases 1.792392, 1.568202, 2.953760, 2.107210, 2.74... #> $ log_deaths 0.3010300, 0.4771213, 1.6020600, 0.3010300, ... ggplot2::ggplot(spatial_br_states, ggplot2::aes(color=cases, size=cases)) + ggplot2::geom_sf()
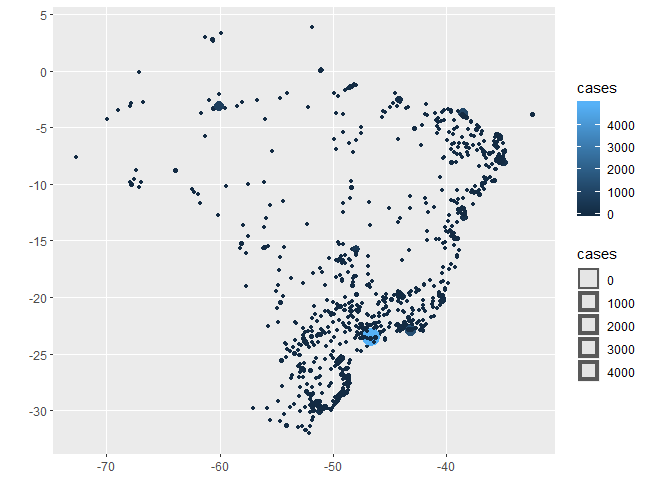
dplyr::glimpse(spatial_br_cities) #> Observations: 908 #> Variables: 7 #> $ date2020-04-08, 2020-04-04, 2020-04-08, 2020-04-08, 2020-04... #> $ city "Abaetetuba", "Abaiara", "Açailândia", "Acrelândia", "Aç... #> $ cases 2, 1, 1, 9, 8, 0, 1, 1, 1, 1, 1, 1, 5, 1, 1, 1, 1, 3, 1,... #> $ deaths 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0,... #> $ geometry POINT (-48.8788 -1.72183), POINT (-39.0416 -7.34588), ... #> $ log_cases 0.3010300, 0.0000000, 0.0000000, 0.9542425, 0.9030900, -... #> $ log_deaths -Inf, -Inf, -Inf, -Inf, -Inf, -Inf, 0, -Inf, 0, -Inf, -I... ggplot2::ggplot(spatial_br_cities, ggplot2::aes(color=cases, size=cases)) + ggplot2::geom_sf()
Data Sources
- States: Ministerio da Saúde
- Cities: brasil.io
- Rio de Janeiro: Secretaria de Saúde - RJ
Links
- Browse source code at
https://github.com/mralbu/coronavirusbrazil/ - Report a bug at
https://github.com/mralbu/coronavirusbrazil/issues
License
- Full license
- MIT + file LICENSE
Developers
- Marcelo Albuquerque
Author, maintainer


